You cannot hide it: Your face tells all about your ID
Last Updated on November 8, 2019 by Joseph Gut – thasso
January 08, 2019 – The interpretation of genetic variants after genome-wide analysis is complex in a selection of heterogeneous disorders which are at the base of a condition termed intellectual disability (ID). Generally, the cause of intellectual disability (ID) is unknown in about one-

third to one-half of cases. About 5% of cases are inherited from a person’s parents. Genetic defects that cause intellectual disability but are not inherited can be caused by accidents or mutations in genetic development. Examples of such accidents are development of an extra chromosome 18 (Trisomy 18) and Down syndrome (Trisomy 21), which is the most common genetic cause. Velocardiofacial syndrome and fetal alcohol spectrum disorders are the two next most common causes. Other conditions, based on genetic aberrations may include Koolen-de-Vries Syndrome, Klinefelter syndrome, Fragile X syndrome, neurofibromatosis, congenital hypothyroidism, Williams syndrome, phenylketonuria (PKU), and Prader–Willi syndrome, Phelan-McDermid syndrome (22q13del), Mowat–Wilson syndrome, genetic ciliopathy, and Siderius type X-linked intellectual disability (OMIM as caused by mutations in the PHF8 gene. In the rarest of cases, abnormalities with the X- or Y-chromosome may also cause disability
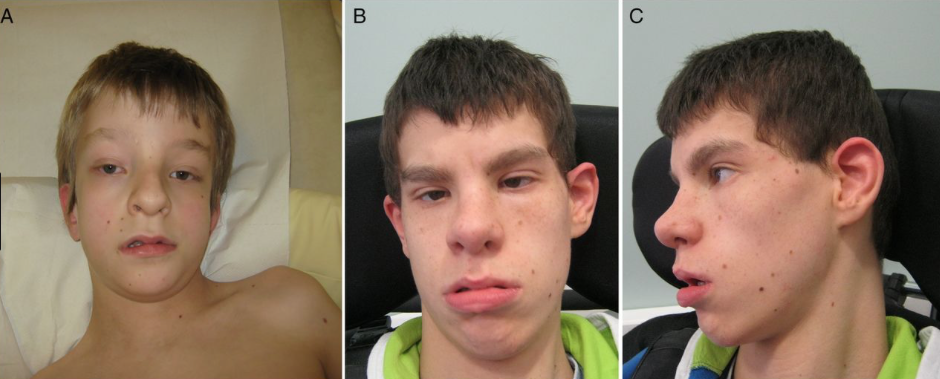
In a very recent study published the Journal Genetics in Medicine (GiM), the authors investigated whether algorithms can be used to detect if a facial gestalt is present for three novel ID syndromes and if these techniques can help interpret variants of uncertain significance. Facial features were extracted from photos of ID patients, more precisely, from patients suffering from Coolen-de-Vries Syndrome, harboring a pathogenic variant in three novel ID genes (PACS1, PPM1D, and PHIP) using algorithms that model human facial dysmorphism, and facial recognition. The resulting features were combined into a hybrid model to compare the three cohorts against a background ID population.
The authors validated their model using images from 71 individuals with Koolen–de Vries syndrome, and were able to show that facial gestalts are present for individuals with a pathogenic variant in PACS1 gene, in PPM1D gene, and also in the PHIP gene. Moreover, two individuals with a de novo missense variant of uncertain significance in the PHIP gene have significant similarity to the expected facial phenotype of PHIP gene-variants bearing patients. Their results show that analysis of facial photos can be used to detect previously unknown facial gestalts for novel ID syndromes, which will facilitate both clinical and molecular diagnosis of rare and novel syndromes.
These are very exciting findings in that they point in the direction that in the future genetic predispositions for certain diseases could be detected by non-invasive face recognition techniques correlating facial phenotypes with the underlying genetic outfit of the concerned individual (patient). It comes then as no surprise, that there exist already at least one company (Face2Gene), which has embarked on the development of a suite of deep phenotyping applications (App’s) that facilitate comprehensive and precise genetic evaluations. Imagine that one day, you are consulting your Selfie, taken on your smartphone, and you can immediately recognize what actually you are suffering from, or what you are going to suffer from later in your life. Bad luck if your Selfie would tell you that you do not belong to the “smartest” of all people.
We may add here that also whole-genome sequencing can be used to diagnose intellectual disability accurately according to a new report in the scientific journal Genome Medicine. Thus, in this report, an astonishingly estimated 1.5 percent of the Swedish population have some sort of intellectual disability, caused in most cases by genetic aberrations, anything from small point mutations (one or a few base pairs) in individual genes or a few base-pair repetitions, to larger structural chromosome mutations comprising one or more genes, including those genes described here. A combination of face recognition and whole genome sequencing may constitute a powerful clinical diagnostic strategy for the diagnosis of intellectual disability.
See here a discussion / explanation on deep phenotyping:


0 Comments on “You cannot hide it: Your face tells all about your ID”